Page Not Found
Page not found. Your pixels are in another canvas. Read more
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Page not found. Your pixels are in another canvas. Read more
This is a page not in th emain menu Read more
Published:
Published:
This is a list of some resources I find useful to conduct data analysis in Life Sciences Research, some of them more specific to the field of Genomics, Transcriptomics, and RNA-seq derived methods. Read more
Published:
I just got my first PhD paper published, which includes the publication of my first software. So I thought it was time to make my own site to organize these and future work. Read more
Published in Frontiers in Physiology, 2015
This review’s aim is to give an overview of what pathway analysis is and how different methods have developed encompassing the same idea of trying to identify biologically meaningful categories that are relevant for the results obtained from Omics experimental data. 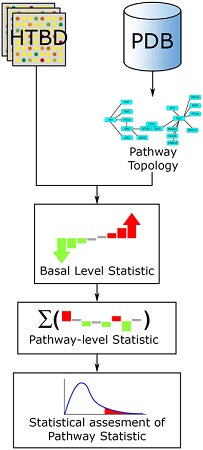
Read more
Recommended citation: García-Campos, M.A., Espinal-Enríquez, J. and Hernández-Lemus, E., 2015. Pathway analysis: state of the art. Frontiers in physiology, 6, p.383. https://www.frontiersin.org/articles/10.3389/fphys.2015.00383
Published in bioRxiv, 2019
In this preprint we present a method to quantify m6a using a m6a sensitive endonuclease, and uncover the mechanistic behaviour of methylation trough a simple and conserved code in cis. 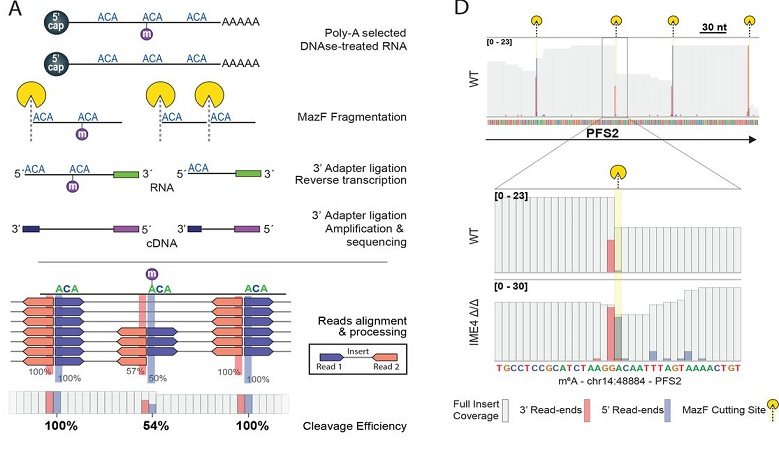
’ Read more
Recommended citation: Garcia-Campos, M.A., Edelheit, S., Toth, U., Shachar, R., Nir, R., Lasman, L., Brandis, A., Hanna, J.H., Rossmanith, W. and Schwartz, S., 2019. Deciphering the "m6A code" via quantitative profiling of m6A at single-nucleotide resolution. BioRxiv, p.571679. https://www.biorxiv.org/content/10.1101/571679v1
Published in Cell, 2019
In this paper we present a method to quantify m6a using an m6a sensitive endonuclease, and uncover the mechanistic behaviour of methylation trough a simple and conserved code in cis.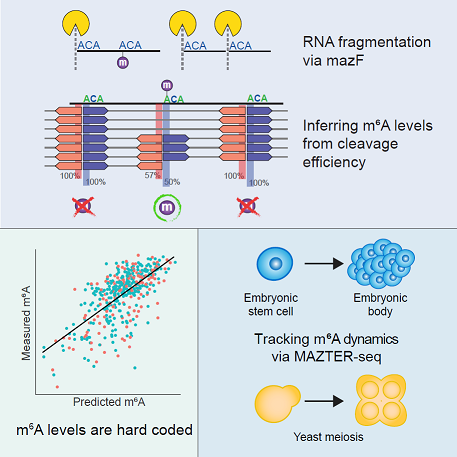
Read more
Recommended citation: Garcia-Campos, M. A. et al. (2019) ‘Deciphering the “m6A Code” via Antibody-Independent Quantitative Profiling’, Cell. Elsevier, 0(0). doi: 10.1016/j.cell.2019.06.013. https://www.cell.com/cell/fulltext/S0092-8674(19)30676-2
Published in Current Microbiology, 2020
In this study we show the recovery and enhancement of the budding yeast’s resistance to oxidative stress by expressing ortholog catalase genes from D. hansenii, a sea yeast. 
Read more
Recommended citation: González, J., Castillo, R., García-Campos, M. A., Noriega-Samaniego, D., Escobar-Sánchez, V., Romero-Aguilar, L., ... & Segal-Kischinevzky, C. (2020). Tolerance to Oxidative Stress in Budding Yeast by Heterologous Expression of Catalases A and T from Debaryomyces hansenii. Current Microbiology, 77(12), 4000-4015. https://link.springer.com/article/10.1007/s00284-020-02237-3
Published in Nature Methods, 2021
Here we develop m6A-seq2, a multiplexed immunoprecipitation-RNA-seq coupled method that measures m6A across the epitranscriptome reducing technical variability, cost, and labor. Allowing for sample-level m6A relative quantitations, orthogonal to mass-spectrometry methods, as well as gene-level quantitations. We found that m6A-seq2 m6A gene-level measurements explains roughly 30% of the variability in the RNA half life on mouse embryonic stem cells. 
Read more
Recommended citation: Dierks, D., Garcia-Campos, M.A., Uzonyi, A., Safra, M., Edelheit, S., Rossi, A., Sideri, T., Varier, R.A., Brandis, A., Stelzer, Y. and van Werven, F., 2021. Multiplexed profiling facilitates robust m6A quantification at site, gene and sample resolution. Nature Methods, pp.1-8. https://www.nature.com/articles/s41592-021-01242-z
Published in bioRxiv, 2023
In this preprint we present txtools, an R package that enables the processing, analysis, and visualization of RNA-seq data at the nucleotide-level resolution, seamlessly integrating alignments to the genome with a transcriptomic representation.
Read more
Recommended citation: Garcia-Campos, M. A., & Schwartz, S. (2023). txtools: an R package facilitating analysis of RNA modifications, structures, and interactions. bioRxiv, 2023-08. https://www.biorxiv.org/content/10.1101/2023.08.24.554738
Published in BMC - Genome Biology, 2024
In this paper we dissect the determinants governing RNA methylation via interspecies and intraspecies hybrids in yeast and mammalian systems, coupled with massively parallel reporter assays and m6A-QTL reanalysis.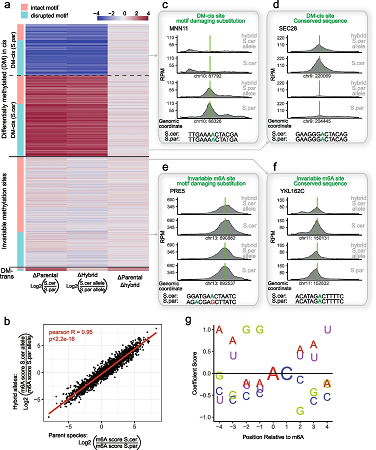
Read more
Recommended citation: Shachar, R., Dierks, D., Garcia-Campos, M. A., Uzonyi, A., Toth, U., Rossmanith, W., & Schwartz, S. (2024). Dissecting the sequence and structural determinants guiding m6A deposition and evolution via inter-and intra-species hybrids. Genome Biology, 25(1), 1-29. https://genomebiology.biomedcentral.com/articles/10.1186/s13059-024-03182-1
Published in Nucleic Acids Research, 2024
We present txtools, an R package that enables the processing, analysis, and visualization of RNA-seq data at the nucleotide-level resolution, seamlessly integrating alignments to the genome with transcriptomic representation. .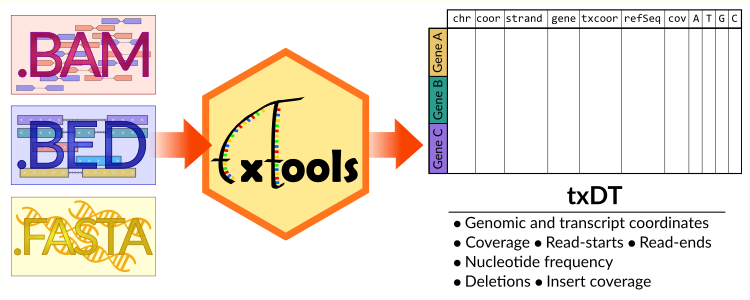
Read more
Recommended citation: Miguel Angel Garcia-Campos, Schraga Schwartz, txtools: an R package facilitating analysis of RNA modifications, structures, and interactions, Nucleic Acids Research, 2024;, gkae203, https://doi.org/10.1093/nar/gkae203. https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkae203/7632932
Published in eLife, 2025
eLife assessment: This study presents a fundamental finding on how levels of m6A levels are controlled, invoking a consolidated model where degradation of modified RNAs in the cytoplasm plays a primary role in shaping m6A patterns and dynamics, rather than needing active regulation by m6A erasers and other related processes. The evidence is compelling through its use of transcriptome-wide data and mechanistic modeling. Relevant for any reader with an interest in RNA metabolism, this new framework consolidates previous observations and highlights the importance of careful experimentation for evaluation m6A levels. .
Read more
Recommended citation: Dierks, D., Shachar, R., Nir, R., Garcia-Campos, M. A., Uzonyi, A., Wiener, D., ... & Schwartz, S. (2025). Passive shaping of intra-and intercellular m6A dynamics via mRNA metabolism. Elife, 13, RP100448. https://elifesciences.org/articles/100448
Published in Cell, 2025
Pan-Mod-seq enables high-throughput profiling of 16 rRNA modifications across diverse species, revealing that while these chemical markers are largely static in mesophiles, they are highly dynamic in hyperthermophiles. Specifically, a conserved m5C-ac4C module is co-induced by heat and serves a critical role in stabilizing ribosome structure at extreme temperatures. Cryo-EM and biophysical analyses confirm that this modification module provides essential thermostability required for hyperthermophilic growth. 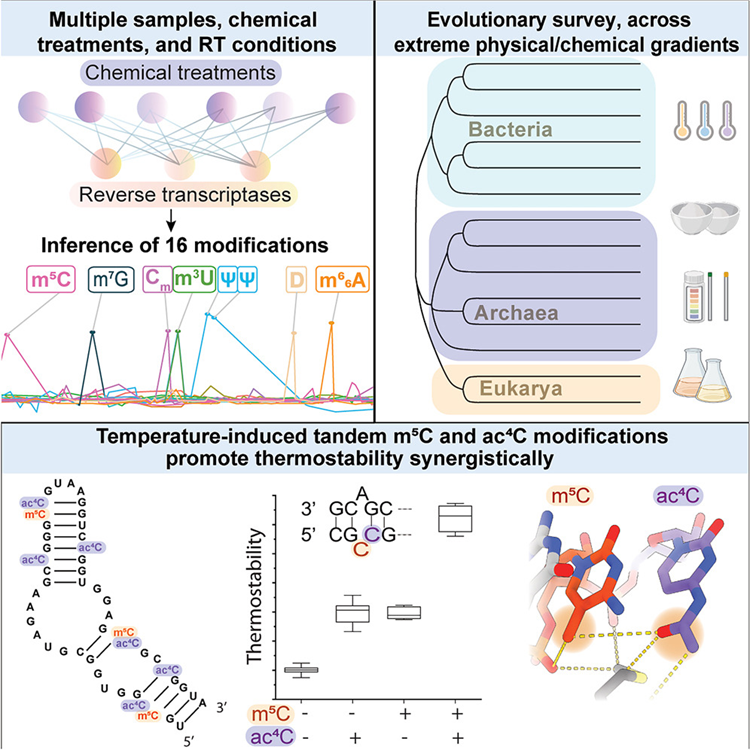
Read more
Recommended citation: Garcia-Campos, M. A., Georgeson, J., Nir, R., Reichelt, R., Fluke, K. A., Matzov, D., ... & Schwartz, S. (2025). Pan-modification profiling facilitates a cross-evolutionary dissection of the thermoregulated ribosomal epitranscriptome. Cell, 188(24), 6825-6844. https://www.cell.com/cell/abstract/S0092-8674(25)01082-7
MAZTER-mine is a computational pipeline to analize MAZTER-seq derived data, a methodology that profiles m6A quantitatively across transcriptomes in a single-base manner. Read more
Published:
A departmental talk on my work on m6A which led to the publication of my first PhD paper. Read more
MSc-PhD course, Weizmann Institute of Science, Feinberg Graduate School, 2017
The course is intended for graduate students in the Life Sciences area. The goal of the course is to enable students to use the R programming language as a tool to extract biological insight from high throughput data. Read more
MSc-PhD course, Weizmann Institute of Science, Feinberg Graduate School, 2018
The course is intended for graduate students in the Life Sciences area. The goal of the course is to enable students to use the R programming language as a tool to extract biological insight from high throughput data. Read more
MSc-PhD course, Weizmann Institute of Science, Feinberg Graduate School, 2019
The course is intended for graduate students in the Life Sciences area. The goal of the course is to enable students to use the R programming language as a tool to extract biological insight from high throughput data. Read more
MSc-PhD course, Weizmann Institute of Science, Feinberg Graduate School, 2020
The course is intended for graduate students in Life Sciences. The goal of the course is to enable students to use the R programming language as a tool to extract biological insight from high throughput data. Read more