Deciphering the “m6A Code” via Antibody-Independent Quantitative Profiling
Published in Cell, 2019
Recommended citation: Garcia-Campos, M. A. et al. (2019) ‘Deciphering the “m6A Code” via Antibody-Independent Quantitative Profiling’, Cell. Elsevier, 0(0). doi: 10.1016/j.cell.2019.06.013. https://www.cell.com/cell/fulltext/S0092-8674(19)30676-2
Summary
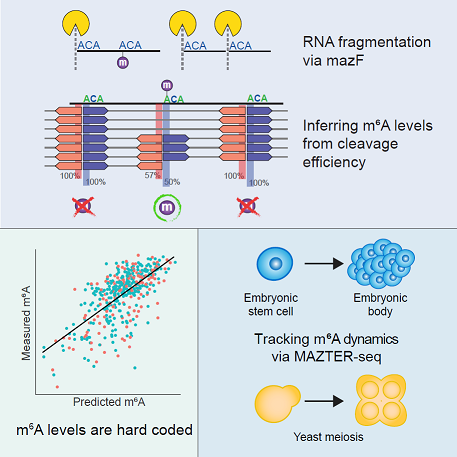
N6-methyladenosine (m 6A) is the most abundant modification on mRNA and is implicated in critical roles in development, physiology, and disease. A major limitation has been the inability to quantify m 6A stoichiometry and the lack of antibody-independent methodologies for interrogating m 6A. Here, we develop MAZTER-seq for systematic quantitative profiling of m6A at single-nucleotide resolution at 16%–25% of expressed sites, building on differential cleavage by an RNase. MAZTER-seq permits validation and de novo discovery of m 6A sites, calibration of the performance of antibody-based approaches, and quantitative tracking of m 6A dynamics in yeast gametogenesis and mammalian differentiation. We discover that m6A stoichiometry is “hard coded” in cis via a simple and predictable code, accounting for 33%–46% of the variability in methylation levels and allowing accurate prediction of m 6A loss and acquisition events across evolution. MAZTER-seq allows quantitative investigation of m 6A regulation in subcellular fractions, diverse cell types, and disease states.

Leave a Comment