Dissecting the sequence and structural determinants guiding m6A deposition and evolution via inter- and intra-species hybrids
Published in BMC - Genome Biology, 2024
Recommended citation: Shachar, R., Dierks, D., Garcia-Campos, M. A., Uzonyi, A., Toth, U., Rossmanith, W., & Schwartz, S. (2024). Dissecting the sequence and structural determinants guiding m6A deposition and evolution via inter-and intra-species hybrids. Genome Biology, 25(1), 1-29. https://genomebiology.biomedcentral.com/articles/10.1186/s13059-024-03182-1
Abstract
Background N6-methyladenosine (m6A) is the most abundant mRNA modification, and controls mRNA stability. m6A distribution varies considerably between and within species. Yet, it is unclear to what extent this variability is driven by changes in genetic sequences (‘cis’) or cellular environments (‘trans’) and via which mechanisms.
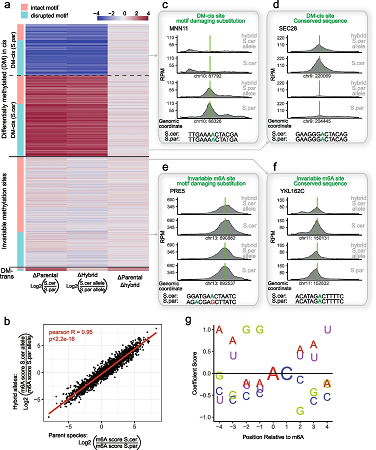
Results Here we dissect the determinants governing RNA methylation via interspecies and intraspecies hybrids in yeast and mammalian systems, coupled with massively parallel reporter assays and m6A-QTL reanalysis. We find that m6A evolution and variability is driven primarily in ‘cis’, via two mechanisms: (1) variations altering m6A consensus motifs, and (2) variation impacting mRNA secondary structure. We establish that mutations impacting RNA structure - even when distant from an m6A consensus motif - causally dictate methylation propensity. Finally, we demonstrate that allele-specific differences in m6A levels lead to allele-specific changes in gene expression.
Conclusions Our findings define the determinants governing m6A evolution and diversity and characterize the consequences thereof on gene expression regulation.

Leave a Comment