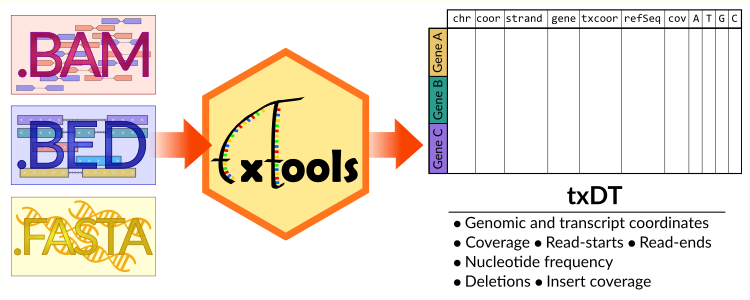
Pan-modification profiling facilitates a cross-evolutionary dissection of the thermoregulated ribosomal epitranscriptome
Published in Cell, 2025
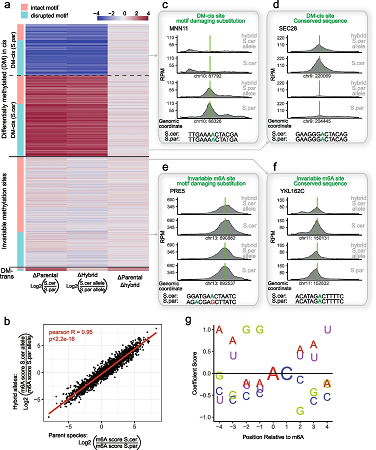
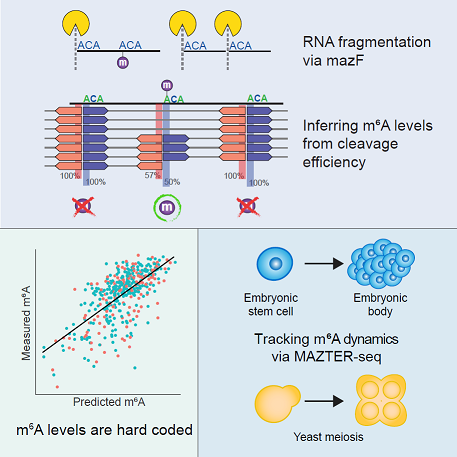
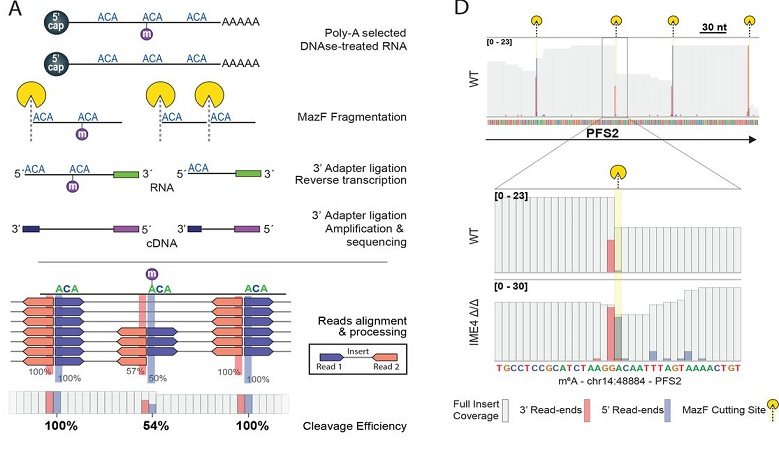
Pan-Mod-seq enables high-throughput profiling of 16 rRNA modifications across diverse species, revealing that while these chemical markers are largely static in mesophiles, they are highly dynamic in hyperthermophiles. Specifically, a conserved m5C-ac4C module is co-induced by heat and serves a critical role in stabilizing ribosome structure at extreme temperatures. Cryo-EM and biophysical analyses confirm that this modification module provides essential thermostability required for hyperthermophilic growth. 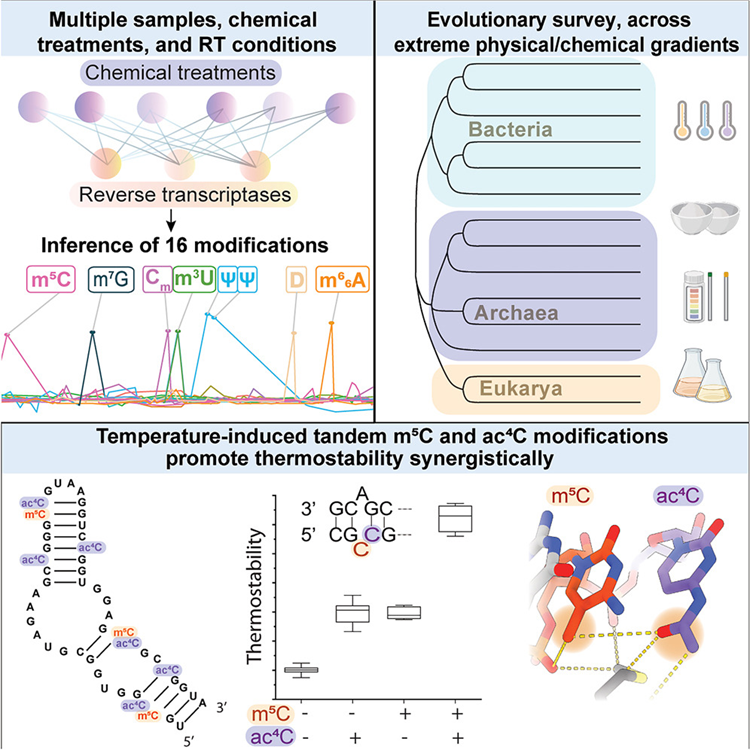
Read more
Recommended citation: Garcia-Campos, M. A., Georgeson, J., Nir, R., Reichelt, R., Fluke, K. A., Matzov, D., ... & Schwartz, S. (2025). Pan-modification profiling facilitates a cross-evolutionary dissection of the thermoregulated ribosomal epitranscriptome. Cell, 188(24), 6825-6844. https://www.cell.com/cell/abstract/S0092-8674(25)01082-7